Knowledge-graph
Rhinitis Knowledge Graph in Traditional Medicine Using NebulaGraph
Author Introduction
Hong Hailan specializes in leveraging large language models (LLMs) to automate the extraction of knowledge from unstructured medical records, construct knowledge graphs in traditional Chinese medicine, and uncover the experiences of renowned TCM practitioners.
Background
Building knowledge graphs presents dual challenges: improving the efficiency of knowledge extraction and reducing human labor costs. To effectively address these issues, this paper builds on the schema layer and explores methods for automated knowledge extraction. The following sections detail the process of constructing a knowledge graph for the treatment of rhinitis by Master Gan Zuwang, a renowned traditional Chinese medicine practitioner.
 The data for this study comes from clinical records of rhinitis patients, with a total of 1,081 detailed records selected for analysis. These valuable records contain various basic patient information, such as name, workplace, gender, age, diagnosis time, disease type, symptom details, diagnostic results, observations of tongue and pulse, treatment records, prescription content, and medication guidance.
The data for this study comes from clinical records of rhinitis patients, with a total of 1,081 detailed records selected for analysis. These valuable records contain various basic patient information, such as name, workplace, gender, age, diagnosis time, disease type, symptom details, diagnostic results, observations of tongue and pulse, treatment records, prescription content, and medication guidance.
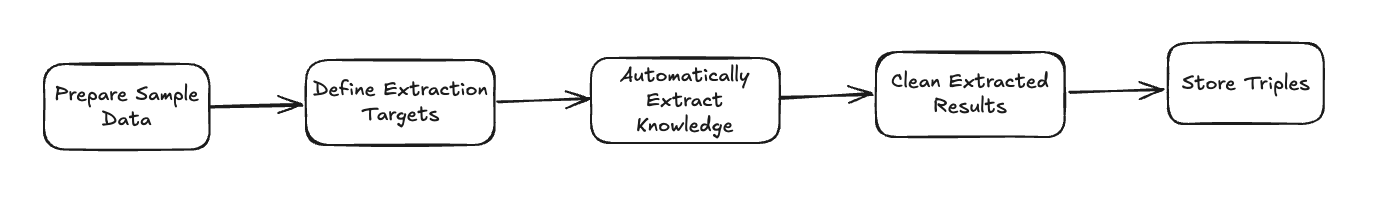
Knowledge Graph Construction Process Diagram
Step 1: Prepare Sample Data
Utilize the 1,081 preprocessed medical records as sample data.
Step 2: Define Extraction Targets
Based on the ontology-defined schema layer, establish extraction targets like diseases, related diseases, symptoms, examinations, tongue appearance, pulse appearance, and compatibility. Define extraction rules for each target type.
Step 3: Automated Knowledge Extraction
Create a prompt template based on the defined extraction targets, using a "demonstration case + relationship list" format to guide large language models in automatically extracting medical record data and generating triples.
The following is a specific Prompt example:
Imagine you are a large language model designed to extract relationships in Traditional Chinese Medicine. Your task is to extract the relationships between symptoms and related diseases for rhinitis patients. Here's an example medical case:
"Rhinitis has been severe for three days, the condition naturally flares up, with excessive nasal discharge, difficulty blowing the nose. Headache. Nasal congestion. Nasal mucosal edema, with secretions. Thin white tongue coating, floating and tight pulse. Prescription: mulberry leaf 6g, peppermint 6g, platycodon root 6g, calamus 6g, spatholobus stem 10g, schizonepeta 6g, angelica dahurica 6g, xanthium fruit 10g, fagopyrum 10g, vitex 10g."
The output format is:[(“rhinitis”, “symptom”, “excessive nasal discharge”), (‘rhinitis’, ‘symptom’, ‘difficulty blowing the nose’), (‘rhinitis’, ‘symptom’, ‘headache’), (‘rhinitis’, ‘symptom’, ‘nasal congestion’), (‘rhinitis’, ‘examination’, ‘nasal mucosal edema’), (‘rhinitis’, ‘examination’, ‘secretions’), (‘rhinitis’, ‘tongue appearance’, ‘thin white tongue coating’), (‘rhinitis’, ‘pulse appearance’, ‘floating and tight pulse’), (‘rhinitis’, ‘combination’, ‘calamus 6g’), (‘rhinitis’, ‘combination’, ‘spatholobus stem 10g’), (‘rhinitis’, ‘combination’, ‘schizonepeta 6g’), (‘rhinitis’, ‘combination’, ‘angelica dahurica 6g’), (‘rhinitis’, ‘combination’, ‘xanthium fruit 10g’), (‘rhinitis’, ‘combination’, ‘fagopyrum 10g’), (‘rhinitis’, ‘combination’, ‘vitex 10g’)]. Follow this format to extract the following medical case in triplet format. The given sentence is: ‘***’. Given the relationship list: [‘symptom’, ‘related disease’, ‘combination’, ‘tongue appearance’, ‘examination’, ‘pulse appearance’], provide the relationships in the list. If they do not exist, output: none.
Step 4: Clean the Extracted Results
Use regular expressions to clean the automatically generated triplet data by removing errors, redundancies, irrelevant, and incomplete triplets. This process ensures the accuracy and reliability of the data.
Step 5: Store the Triplets
Store the final extracted triplet data in the NebulaGraph database.
This paper directly extracts diseases and Chinese herbs into corresponding triplets to ensure the reliability of the knowledge extraction process using large language models. During the writing process, these triplets are further decomposed to ensure the rationality of the knowledge graph. Additionally, the entities and attributes of Chinese herbs are jointly extracted during the knowledge extraction. For example, the dosage of Fagopyrum 10g in a rhinitis patient’s prescription is extracted as ('rhinitis', 'combination', 'fagopyrum 10g').
Before storing the triplets, this paper uses regular expressions to extract the entities and their corresponding attributes. For instance, ('rhinitis', 'combination', 'fagopyrum 10g') is decomposed into ('rhinitis', 'combination', 'Fagopyrum') and ('fagopyrum', 'dosage', '10g'), as demonstrated in the constructed knowledge graph.
Using NebulaGraph to build a knowledge graph
Why NebulaGraph?NebulaGraph is a high-performance and user-friendly graph database that offers researchers a new way to explore and analyze data thanks to its outstanding features and broad applications. It comes with a graph query language called nGQL, which provides users with a rich set of graph operation functions. Whether for basic CRUD operations on nodes and edges or complex graph algorithms and path queries, nGQL handles them effortlessly. This flexibility and robust functionality make NebulaGraph an excellent choice for handling complex network structures and maximizing the value of medical case data.
Specific steps:
Algorithm pseudocode
Construct prompt templates, related cases, and relationship lists
Write functions to connect to NebulaGraph
for i := 0 to N do // traverse medical case data
Generate questions by combining data and templates
Pass the questions to the LLM to get answers
Clean the answers and remove unnecessary triples
Build the IDs for patients, examinations, symptoms, and prescriptions in the current case
for j := 0 to M do // iterate through the triple list
if relation'=="combination”:
Determine whether the Chinese herb in the triplet contains a dosage unit attribute. If it does, extract the attribute and store the extracted result in the knowledge graph.
Else: Directly write the extracted results into the knowledge graph.
end for
Experiment
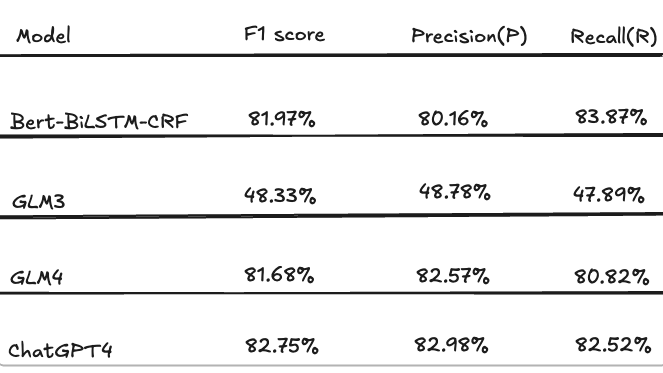
This study utilizes a dataset of 353 pre-prepared medical cases as the foundation for knowledge extraction experiments using large language models (LLMs). In the experiment, other LLMs are employed for data annotation, followed by manual verification and correction to ensure the accuracy and reliability of the annotated data. This process serves to evaluate the knowledge extraction performance of each model. The Bert-BiLSTM-CRF model is selected as the baseline for joint entity and relationship extraction, and the results of automatic extraction using the Bert-BiLSTM-CRF model are compared with those obtained from large language models, primarily GLM3, GLM4, and ChatGPT-4. The evaluation metrics include F1 score, precision, and recall, with the experimental results presented in the table.
Knowledge Graph Display
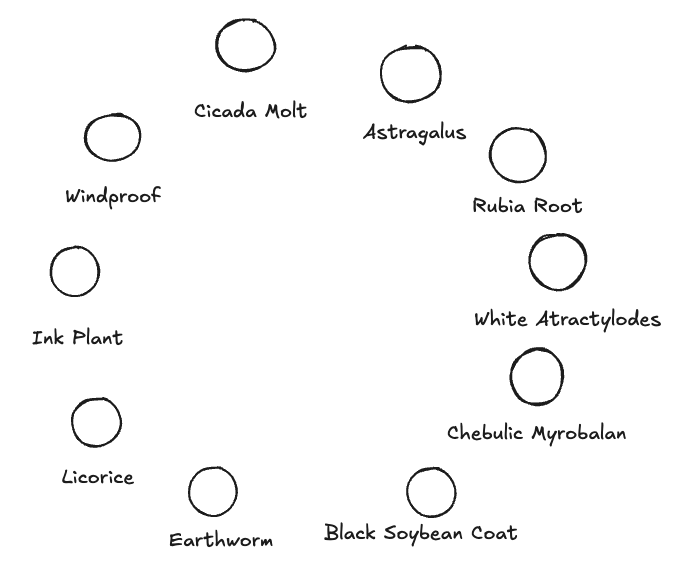
This study leverages the constructed knowledge graph to analyze and mine medication patterns used by a master physician in treating rhinitis, such as identifying the top 10 drugs most frequently prescribed for severe sneezing symptoms. The following Cypher query is executed to achieve this:
MATCH (v:Patient)-[:HAS_SYMPTOM]->()-[:SPECIFIC_SYMPTOM]->(v1)
WHERE id(v1) = 'severe sneezing'
MATCH (v:Patient)-[:PRESCRIBED]->(v3)-[e2:COMPONENT_OF]->(v4)
RETURN v4, count(v4) AS cnt
ORDER BY cnt DESC
LIMIT 10;
This query identifies patients exhibiting severe sneezing symptoms, retrieves their treatment prescriptions, and sorts the drugs by frequency of occurrence, returning the top 10 most frequently prescribed drugs. The top 10 drugs are: Cicada Skin, Windproof, Ink Plant, Licorice, Earthworm, Black Soybean Coat, Chebulic Myrobalan, White Atractylodes, Rubia Root, and Astragalus.